Research Article / Open Access
DOI: 10.31488/jjm.161
Proposal: Search for Small Chemical Compounds Interacting with Bacterial Elongation Factor EF-Tu in its Role in a Bacterial Cytoskeleton -a New Class of Antibiotics?
Prof.Dr.Frank Mayer*
Institute of Microbiology and Genetics, Georg-August-University Goettingen, Germany
*Corresponding author: Prof.Dr.Frank Mayer, retired from Institute of Microbiology and Genetics, Jaspersstrasse 2 O-7/521 69126 Heidelberg, Germany, Tel: +49 6221 388 521,E-mail:fmayer12@gmx.de
Keywords: bacterial cytoskeleton, bacterial elongation factor ef-tu;protein with three domains, truncated protein, competitive inhibition, induced bacterial lysis, micropeptides, small chemical compounds, chemical libraries, antibiotics
Introduction
After the discovery of the existence of bacterial cytoskeletons [1-6] it was described that the bacterial elongation factor EF-Tu does not only have a function in protein synthesis, but also as a component of one of the bacterial cytoskeletons [3,4]. It was also speculated that, in evolution, the primordial bacterial cytoskeleton had EF-Tu as major component [4,6].
Experiments with “truncated“ EF-Tu proteins revealed that, in their presence in the bacterial cell, the cells in growing cultures did end growth due to the fact that their newly formed cytoskeletons had defects. Their cytoskeletons were weakened by integration of truncated EF-Tu in the growing cytoskeleton. Truncated EF-Tu molecules did block, by competitive inhibition with intact EF-Tu molecules for the binding sites in growing cytoskeletons, the binding sites for intact EF-Tu (Figures 1-3).
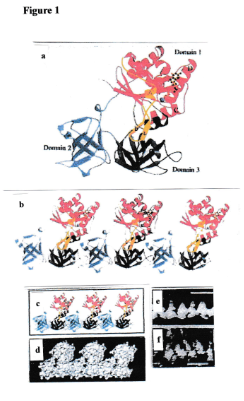
Figure 1.. Conformation of elongation factor EF-Tu and „chain“ formation by polymerization. (a) Ribbon plot on the basis of x-ray crystallography [10]. (b) Chain formation by polymerization, Domain 2 fits into Domain 3 of a neighbouring particle [6]. (c,d) Two kinds of presentation of a chain [6]. (e) Ultrastructure of a chain isolated from a lysed bacterium [6]. (f) Ultrastructure of a chain created by in vitro polymerization of individual EF-Tu proteins [6]. (e) Note the convincing similarity of the structural organization depicted in and (f) with the structural organization shown in d.
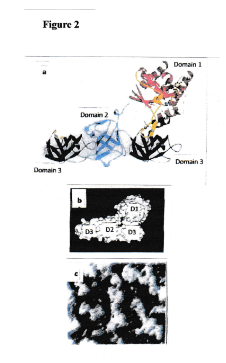
Figure 2.Inhibition of chain formation by insertion of truncated EF-Tu into a growing chain [6]. (a) A „truncated“ EF-Tu protein (consisting only of domain 3) is bound to domain 2 of a full-size EF-Tu protein at the end of a chain. The truncated molecule has occupied the binding site on domain 2 of a full-size EF-Tu molecule. Hence, further chain formation is no longer possible. (b) Image of a model of this complex. (c) Electron micrograph of a sample of EF-Tu molecules with bound truncated EF-Tu molecules (only domain 3). Note short and ultrashort complexes. No chains visible. A situation depicted in Figure 3, created by in vitro experiments, is expected when antibiotics simulating a truncated EF-Tu molecule are administered
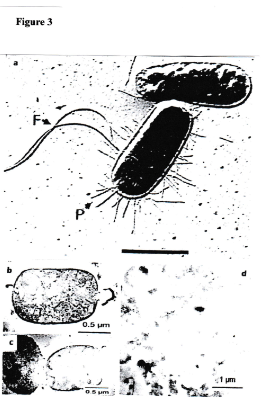
Figure 3.Intact bacterial cells and lysed bacteria prior to and after administration of an antibacterial agens [6]. (a) Intact bacterial cells (Escherichia coli with flagella and fimbriae). (b,c,d) Bacterial cell remnants after administration of an antibacterial agent (antibiotic). Note several states of lysis. These cells are dead. The human immune system takes care of the final degradation of the remnants.
Results, Discussion, Conclusions
On the basis of the findings described above, an idea was born: micropeptides or other compounds simulating the binding sites of the EF-Tu proteins forming chains (the elements of cytoskeletons in bacteria), applied as antibiotics, could be found. Synthesis of truncated Ef-Tu protein molecules with such a function was not envisaged because their size may be too large for transport into the bacterial cell. Nevertheless, a world patent [7] was awarded that protects synthesis and application of micropeptides that simulate the binding sites of EF-Tu involved in chain formation.
As mentioned above, transport of compounds into the interior of the bacterial cell, acting as antibiotics as described , is the obvious prerequisite for this kind of antibiotic function.
It can be envisaged that small compounds (not necessarily micropeptides, but other classes of molecules) may be found that replace micropeptides and can be transported into bacterial cells. A search will be necessary in chemical libraries [8,9]. Below, a respective procedure will be described.
Note: this is a proposal; therefore, no results can be communicated
Proposed Procedure
First, a source for a sufficient amount of EF-Tu (individual molecules, not organiszed in chains) has to be found. EF-Tu is a highly conserved protein [11]. The DNA sequence and the amino acid sequence of this three-domain-protein of very different strains of bacteria was compared. It turned out that only very minor differences do exist. Therefore, for the experiments described above and below it is not very important to know from which bacterial strain the EF-Tu sample is obtained.
The next steps in the procedure should be performed as follows. First, a size distribution of particles in the particle sample is investigated by an appropiate technique. The curve can be expected to have one peak, indicating that the EF-Tu molecules are not organized in chains, but are individual unbound molecules. This result can be expected when conditions (pH, salts) are fulfilled that avoid chain formation. When conditions are fulfilled that are favourable for chain formation (pH, salts), the peaks that can be awaited are: a peak for individual EF-Tu molecules (not all molecules in the sample are organized in chains), and several peaks for chains of different length. Now, in each of the wells of a mikrotiter plate an aliquot of the buffer solution is given which is known to favour the formation of chains. EF-Tu is not yet added. Now, aliquots of solutions of various “candidate“ compounds are added. It might be helpful to take a selection of “candidate’’ compounds that are known to bind to proteins. Now aliquots of the sample containing individual EF-Tu molecules are added. After an appropriate time which is needed for chain formation, the contents of the different wells is investigated regarding size distribution of „particles“ in the wells. Those wells will be interesting where no chain formation has occurred. This would indicate not only that the added “candidate“ compound has bound to the EF-Tu protein. Much more important would be that the „candidate“ compound has bound to the sites on the EF-Tu protein that are responsible for chain formation. This indicates inhibition of chain formation. This result is actually the aim of the experiment.
The amino acid sequence and conformation of the exposed binding sites in domains 2 and 3 of the EF-Tu protein are known from x-ray crystallography [10]. Hence, one could use this knowledge for optimization experiments regarding the structure and function of the “candidate“ compound.
A very important aspect of this laboratory experiment would be to find ways for the transport of the “candidate“ protein into the bacterial cell. Only then, the “candidate“compound would really be an antibacterial agent called antibiotic.A name for such an antibiotic could be “Nanocytocin’’. Such a name, especially the “nano’’, would indicate that this antibacterial agent is derived from a structural element of nanometer size in the bacterial cell.
References
1. Antranikian G, Herzberg C, Mayer F, et al. Changes in the cell envelope structure of Clostridium sp. Strain EM1 during massive production of alpha-amylase and pullulanase. FEMS MicrobiolLett. 1987; 41: 193-197.
2. Mayer F, Vogt B, Poc C. Immunoelectron microscopic studies indicate the existence of a cell shape preserving cytoskeleton in prokaryotes. Naturwissenschaften.1998; 85:278-282
3. Hegermann J, Herrmann R, Mayer F. Cytoskeletal elements in the bacterium Mycoplasma pneumoniae. Naturwissenschaften. 2002;89: 453-458
4. Madkour MHF, Mayer F. Intracellular cytoskeletal elements and cytoskeletons in bacteria. Science Progress. 2007;90/2: 59-88
5. Mayer F. Cytoskeletons in Prokaryotes – Status report and hypotheses. Cell Biol Internat. 2003;27: 429-438
6. Mayer F. Role of Elongation Factor EF-Tu in bacterial cytoskeletons – Mini Review and Update. Swift JMed MedSci. 2015; 1 (2): 06-11.
7. Mayer F. Worldpatent WO 2002087554 A2. EF-Tu binding substances as antibacterial agens. 2002.
8. Rudolph MG. Leitmolekuelsuche mit DNA-codierten Bibliotheken. BIOSpektrum 02.17, 23.Jahrgang. 2002; 142-145
9. Franzini RM, neri D, Scheuermann J. DNA-encoded libraries: advancing beyond conventional small-molecule libraries. Acc Chem Res. 2014; 47: 1247-1255.
10. Song H, Parsons MR, Roswell S, et al. Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation ar 2.05 A resolution. J Mol Biol. 1999;285: 1245-1256
11. "https://en.wikipedia.org/wiki/EF-Tu" (in the introduction to the chapter and in “Evolution’’)
Received: November 22,2019;
Accepted:December 26, 2019;
Published:December 31, 2019.
To cite this article : F Mayer. Proposal: Search for Small Chemical Compounds Interacting with Bacterial Elongation Factor EF-Tu in its Role in a Bacterial Cytoskeleton - a New Class of Antibiotics?. Japan Journal of Medicine. 2020;3:3.
© F Mayer. 2020.
